
Multitasking of Hsp70 chaperone in the biogenesis of bacterial functional amyloids.
Shinya Sugimoto, Ken-ichi Arita-Morioka, Akari Terao, Kunitoshi Yamanaka, Teru Ogura, Yoshimitsu Mizunoe.
Communications Biology, 1, Article number: 52 (2018)
doi.org/10.1038/s42003-018-0056-0
Biofilms are intricate communities of microorganisms embedded in a self-produced matrix of extracellular polymer, which provides microbes survival advantages in stressful environments and can cause chronic infections in humans. Curli are functional amyloids that assemble on the extracellular surface of enteric bacteria such as Escherichia coli during biofilm development and colonization. The molecular chaperone DnaK, a bacterial Hsp70 homolog, promotes curli biogenesis via unknown mechanism(s). Here we show that DnaK increases the expression of CsgA and CsgB—the major and minor structural components of curli, respectively—via a quantity and quality control of RpoS, a stationary phase-specific alternative sigma factor regulating bacterial transcription, and CsgD, the master transcriptional regulator of curli formation. DnaK also keeps CsgA and CsgB in a translocation-competent state by binding to their signal peptides prone to aggregation. Our findings suggest that DnaK controls the homeostasis of curli biogenesis at multiple stages to organize the biofilm matrix.
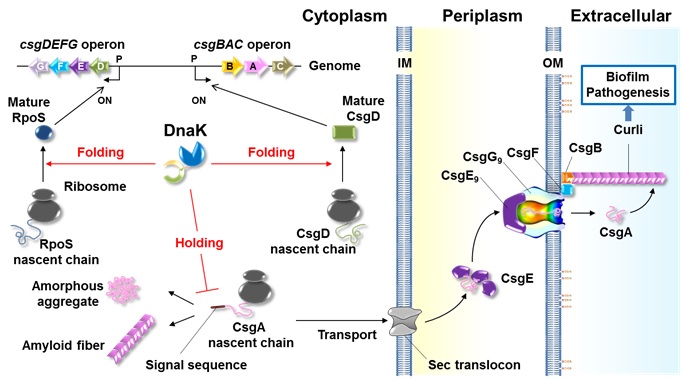
Figure. Model for DnaK functions in curli biogenesis.
DnaK regulates quantity and quality of RpoS, promoting expression of the csgDEFG operon. DnaK also assists de novo folding of CsgD, which activates the csgBAC operon. DnaK recognizes N-terminal end of signal peptide of CsgA and CsgB and keeps their transport competent state by preventing aggregation (holding), which likely accelerates successful translocation of these amyloidogenic proteins into the periplasm.