
Hidekazu Naganuma, Koichiro Miike, Tomoko Ohmori, Shunsuke Tanigawa, Takumi Ichikawa, Mariko Yamane, Masatoshi Eto, Hitoshi Niwa, Akio Kobayashi, Ryuichi Nishinakamura. Molecular detection of maturation stages in the developing kidney. Developmental Biology, in press (2020)
https://www.sciencedirect.com/science/article/pii/S0012160620302943
Recent advances in stem cell biology have enabled the generation of kidney organoids in vitro, and further maturation of these organoids is observed after experimental transplantation. However, the current organoids remain immature and their precise maturation stages are difficult to determine because of limited information on developmental stage-dependent gene expressions in the kidney in vivo. To establish relevant molecular coordinates, we performed single-cell RNA sequencing (scRNA-seq) on developing kidneys at different stages in the mouse. By selecting genes that exhibited upregulation at neonates (P0) compared with embryonic day (E) 15.5 as well as cell lineage-specific expression, we generated gene lists correlated with developmental stages in individual cell lineages. Application of these lists to transplanted embryonic kidneys revealed that most cell types, other than the collecting ducts, exhibited similar maturation to kidneys at the neonatal stage in vivo, revealing non-synchronous maturation across the cell lineages. Thus, our scRNA-seq data can serve as useful molecular coordinates to assess the maturation of developing kidneys and eventually of kidney organoids.
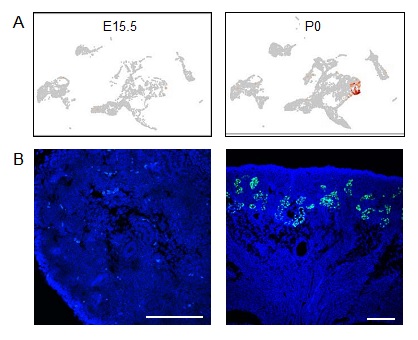
Figure. An example of stage-dependent gene during kidney maturation.
(A) Stage-specific scRNA-seq plots of Cyp27b1 in E15.5 and neonatal (P0) kidneys.
(B) In situ hybridization of Cyp27b1 in E15.5 and P0 kidneys. Scale bar: 200 µm.